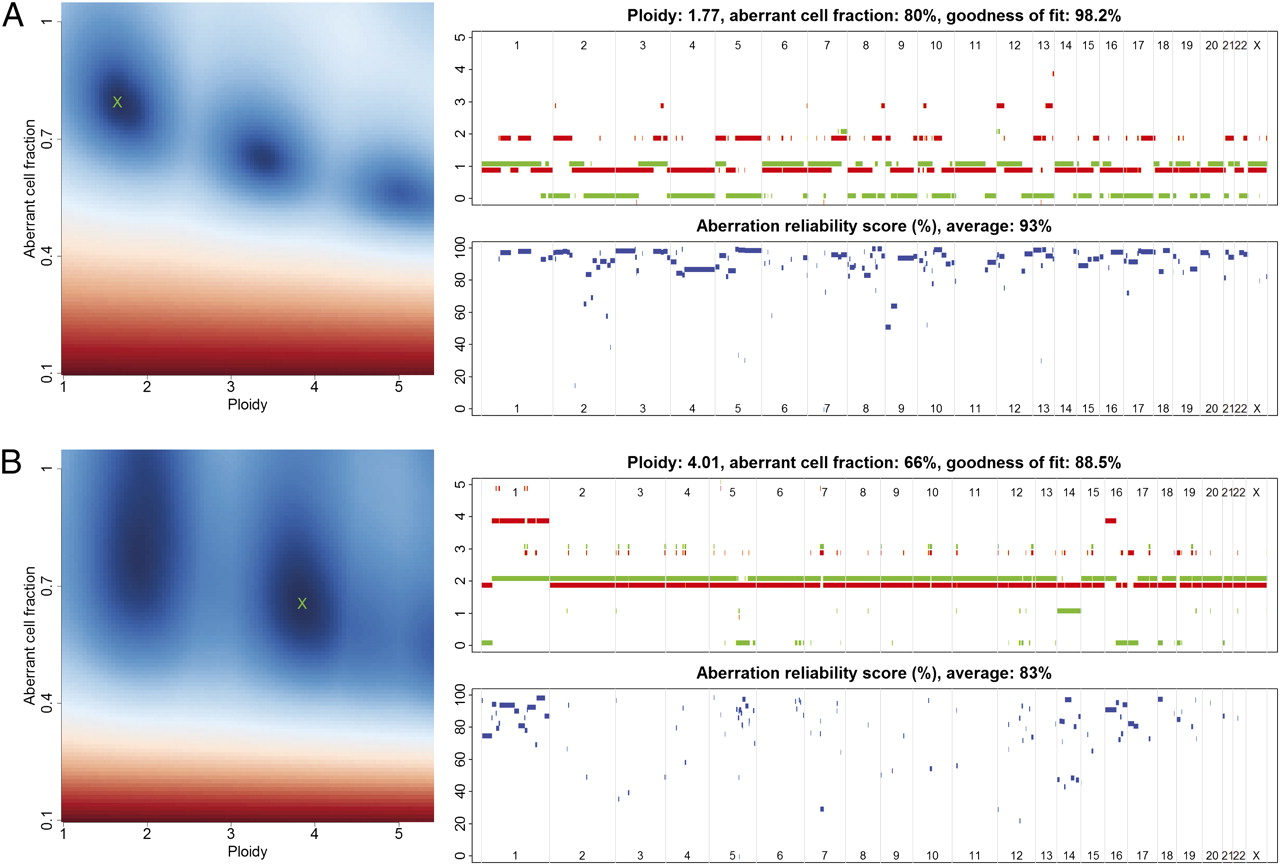
GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low-coverage whole genome sequencing data
By A Mystery Man Writer
Last updated 22 Sept 2024
Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE
RNAseqCNV: analysis of large-scale copy number variations from RNA-seq data
Comprehensive Assessment of Somatic Copy Number Variation Calling Using Next-Generation Sequencing Data
PDF) OTSUCNV: an adaptive segmentation and OTSU-based anomaly classification method for CNV detection using NGS data
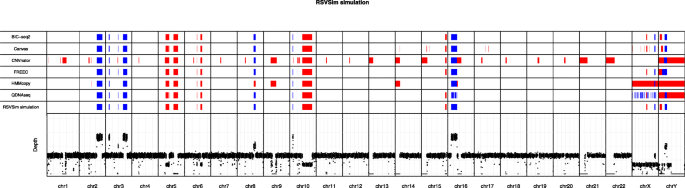
Evaluation of tools for identifying large copy number variations from ultra- low-coverage whole-genome sequencing data, BMC Genomics
GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low-coverage whole genome sequencing data
OUH - Cancer Genome Variation
Assessing Copy Number Alterations in Targeted, Amplicon-Based Next-Generation Sequencing Data - ScienceDirect
QIAGEN Omicsoft Copy Number Variation Analysis tutorial
GitHub - AdelmanLab/GetGeneAnnotation_GGA
Recommended for you
 Understanding and increasing coverage : Knowledge Base14 Jul 2023
Understanding and increasing coverage : Knowledge Base14 Jul 2023 The variables for NGS experiments: coverage, read length, multiplexing14 Jul 2023
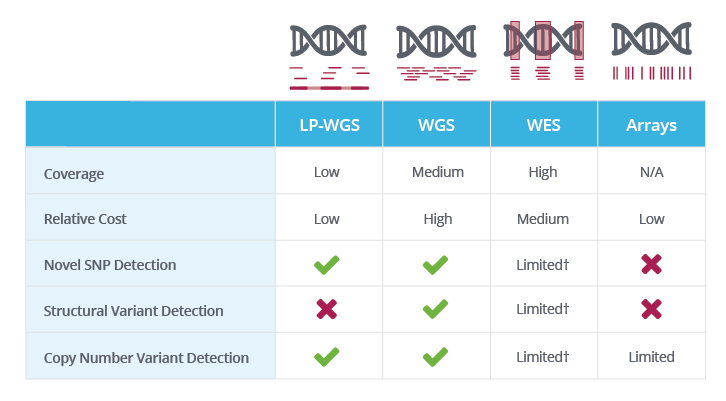
The variables for NGS experiments: coverage, read length, multiplexing14 Jul 2023- Low-Pass Whole Genome Sequencing14 Jul 2023
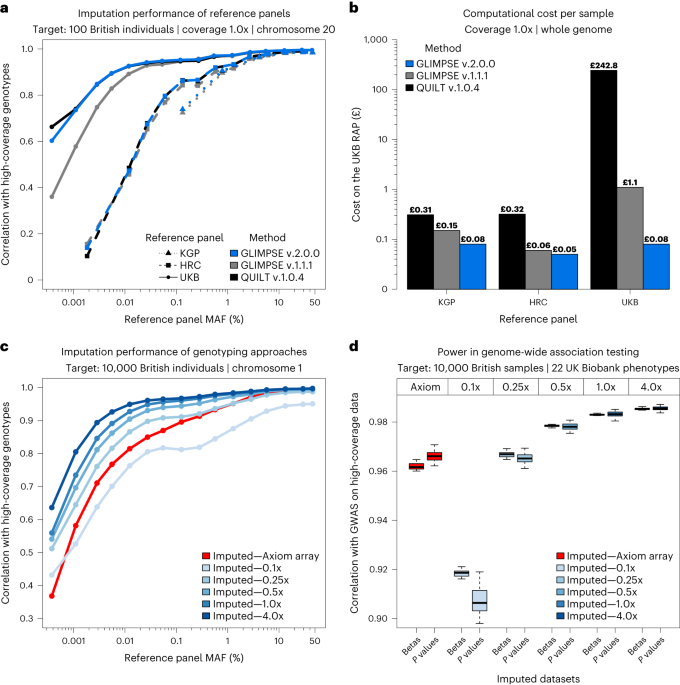 Imputation of low-coverage sequencing data from 150,119 UK Biobank genomes14 Jul 2023
Imputation of low-coverage sequencing data from 150,119 UK Biobank genomes14 Jul 2023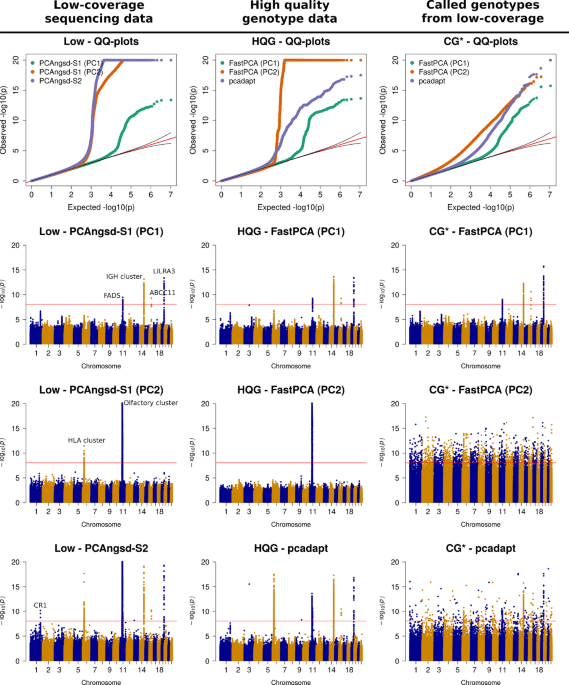 Detecting selection in low-coverage high-throughput sequencing data using principal component analysis, BMC Bioinformatics14 Jul 2023
Detecting selection in low-coverage high-throughput sequencing data using principal component analysis, BMC Bioinformatics14 Jul 2023 Low-Signal Icons - Free SVG & PNG Low-Signal Images - Noun Project14 Jul 2023
Low-Signal Icons - Free SVG & PNG Low-Signal Images - Noun Project14 Jul 2023 Antarctic sea ice hits 'record-smashing' low coverage area, new data show, Climate Crisis News14 Jul 2023
Antarctic sea ice hits 'record-smashing' low coverage area, new data show, Climate Crisis News14 Jul 2023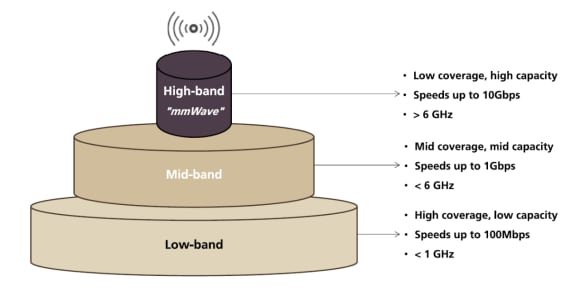 5G coverage vs. capacity14 Jul 2023
5G coverage vs. capacity14 Jul 2023 Cotton/Hosiery non-padded lower Back/Backless single-Hook ,Low Coverage,V-shap bra for women14 Jul 2023
Cotton/Hosiery non-padded lower Back/Backless single-Hook ,Low Coverage,V-shap bra for women14 Jul 2023 Coverage-Driven Verification Isn't Complete Without Low-Power Metrics14 Jul 2023
Coverage-Driven Verification Isn't Complete Without Low-Power Metrics14 Jul 2023
You may also like
 Your Forgiveness Coupons Common-Sense Spirituality14 Jul 2023
Your Forgiveness Coupons Common-Sense Spirituality14 Jul 2023 Zara Leather Pants (Brand New), Women's Fashion, Bottoms, Other14 Jul 2023
Zara Leather Pants (Brand New), Women's Fashion, Bottoms, Other14 Jul 2023 Lacey Bikini in Lilac By Uye Surana - XS-5X - Gigi's - Toronto14 Jul 2023
Lacey Bikini in Lilac By Uye Surana - XS-5X - Gigi's - Toronto14 Jul 2023 Snowboard Full Cushion Mountain Geo Pattern OTC Socks14 Jul 2023
Snowboard Full Cushion Mountain Geo Pattern OTC Socks14 Jul 2023 Victoria's Secret14 Jul 2023
Victoria's Secret14 Jul 2023- Hands Free Pumping Bra, Momcozy … curated on LTK14 Jul 2023
 Chaco Classic Flip-Flops (For Men) - Save 27%14 Jul 2023
Chaco Classic Flip-Flops (For Men) - Save 27%14 Jul 2023 Dettol Hand Soap Original 250ml - Guardian Online Malaysia14 Jul 2023
Dettol Hand Soap Original 250ml - Guardian Online Malaysia14 Jul 2023 Soma, Intimates & Sleepwear, Nwt Nwot Lot Of Victorias Secret Soma Bras Size 36c 38b14 Jul 2023
Soma, Intimates & Sleepwear, Nwt Nwot Lot Of Victorias Secret Soma Bras Size 36c 38b14 Jul 2023 discounted clearance BRAND-NEW Nike Pro Leggings14 Jul 2023
discounted clearance BRAND-NEW Nike Pro Leggings14 Jul 2023